- PRODUCTS
-
UP.SIGHT™ 2nd GenNEW
Optimized For Proof Of Monoclonality, Colony Tracking, Confluency, & Titer Measurement -
F.SIGHT™ 2.0
Optimized For Rapid Dispensing of Fluorescent Cells -
C.SIGHT™ 2.0
Optimized for Powerful Dispensing of Unlabeled Cells -
B.SIGHT™
Optimized For Rapid Microbial Single-Cell Isolation and Cultivation -
F.SIGHT™ OMICS
Optimized For Single-Cell-Omics -
F.SIGHT™
Optimized For Affordability And Flexibility -
C.SIGHT™
Optimized for Affordable Cell Line Development -
Compare Products
Decide which one is right for you - Help Me Choose
-
UP.SIGHT™ 2nd GenNEW
- APPLICATIONS
- RESOURCES HUB
- COMPANY
- SHOP
NEW PROTOCOL FOR FASTER AND MORE RELIABLE RNA COUNTING
Postdoctoral researcher Christoph Ziegenhain has developed a new protocol for faster and more reliable RNA counting. Based at the Sandberg Lab at the Karolinska Institute in Stockholm, Sweden, Ziegenhain focuses primarily on single-cell quantitative transcriptomics methods. His latest paper, published in Nature Methods, is entitled “Molecular spikes: a gold standard for single-cell RNA counting” and describes how he and his colleagues successfully used CYTENA’s F.SIGHT™ OMICS to develop a new quality control procedure for RNA counting in experiments.
scRNA-seq is commonly used to infer cellular identities in healthy and diseased tissues. Many scRNA-seq methods rely on counting strategies to compensate for amplification biases when quantifying RNA molecules. For this, a unique molecular identifier (UMI) typically consisting of a random short sequence is introduced during reverse transcription so that each RNA molecule is uniquely barcoded in the process. However, this approach is subject to errors during amplification and sequencing, and carries a risk of failures which can potentially result in artifactual RNA counts despite the use of UMIs. Therefore, there is a clear need for systematic quality control and benchmarking of the counting process in new scRNA-seq methods and modifications to established protocols.
NEW PROTOCOL FOR FASTER AND MORE RELIABLE RNA COUNTING
Postdoctoral researcher Christoph Ziegenhain has developed a new protocol for faster and more reliable RNA counting. Based at the Sandberg Lab at the Karolinska Institute in Stockholm, Sweden, Ziegenhain focuses primarily on single-cell quantitative transcriptomics methods. His latest paper, published in Nature Methods, is entitled “Molecular spikes: a gold standard for single-cell RNA counting” and describes how he and his colleagues successfully used CYTENA’s F.SIGHT™ OMICS to develop a new quality control procedure for RNA counting in experiments.
scRNA-seq is commonly used to infer cellular identities in healthy and diseased tissues. Many scRNA-seq methods rely on counting strategies to compensate for amplification biases when quantifying RNA molecules. For this, a unique molecular identifier (UMI) typically consisting of a random short sequence is introduced during reverse transcription so that each RNA molecule is uniquely barcoded in the process. However, this approach is subject to errors during amplification and sequencing, and carries a risk of failures which can potentially result in artifactual RNA counts despite the use of UMIs. Therefore, there is a clear need for systematic quality control and benchmarking of the counting process in new scRNA-seq methods and modifications to established protocols.
HOW TO OVERCOME THE PROBLEM
Ziegenhain and his colleagues proposed a new approach in which they introduced RNA spike-ins containing built-in UMIs to identify and quantify artifactual RNA counting. After validating the randomness and complexity of these spike-ins, the authors verified the counting accuracy of the SMART-seq3 protocol and also showed that this approach can correctly diagnose inaccurate counting introduced by suboptimal reaction conditions. For instance, the Sandberg lab team explained that their new tool revealed “completely artificial” results from a variant of the SCRB-seq protocol. This effect is caused by preferential priming of UMI-containing oligo-dT primers during PCR amplification, thus introducing false UMIs every PCR cycle and resulting in an overcount of those UMIs and a false read.
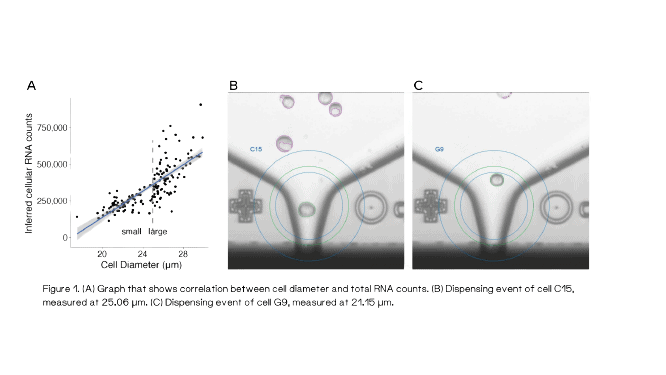
To further validate the spike-UMI approach, the authors then took advantage of the F.SIGHT OMICS and its unique feature to capture nozzle brightfield images for each isolated single cell along with the corresponding cell diameter. Single HEK293FT cells were precisely deposited into 384-well plates containing only 0.3 μL lysis buffer (+3 μL hydrophobic overlay) and then processed according to the Smart-seq3xpress protocol. By calculating the ratio of spike-in molecules to the cells’ endogenous RNA molecules, the authors could infer the absolute mRNA content of each cell which correlated to its diameter (Figure 1). The clear is normally masked by varying levels of sequencing saturation, which could successfully be corrected using the spike-in approach.
CONCLUSION
The study clearly showed that the molecular spikes can be applied in scRNA-seq experiments to diagnose and quantify artifactual RNA counting. Moreover, the authors used the F.SIGHT OMICS to truly play out the unique strengths of the device: highly precise, fast and efficient single-cell dispensing, and the simultaneous recording of true image data on each single cell dispensed. Using this comprehensive image data, researchers can integrate cell diameter in their analysis of scRNA-seq datasets, contributing to the complexity, accuracy and reliability of the results.
The study clearly showed that the molecular spikes can be applied in scRNA-seq experiments to diagnose and quantify artifactual RNA counting. Moreover, the authors used the F.SIGHT OMICS to truly play out the unique strengths of the device: highly precise, fast and efficient single-cell dispensing, and the simultaneous recording of true image data on each single cell dispensed. Using this comprehensive image data, researchers can integrate cell diameter in their analysis of scRNA-seq datasets, contributing to the complexity, accuracy and reliability of the results.
References:
Ziegenhain, C., Hendriks, GJ., Hagemann-Jensen, M. et al. Molecular spikes: a gold standard for single-cell RNA counting. Nat Methods 19, 560–566 (2022). https://doi.org/10.1038/s41592-022-01446-x
Products used:
F.SIGHT™ OMICS
Aim:
Capture nozzle brightfield images for each isolated single cell along with the corresponding cell diameter to validate the spike-UMI approach.

HOW TO OVERCOME THE PROBLEM
Ziegenhain and his colleagues proposed a new approach in which they introduced RNA spike-ins containing built-in UMIs to identify and quantify artifactual RNA counting. After validating the randomness and complexity of these spike-ins, the authors verified the counting accuracy of the SMART-seq3 protocol and also showed that this approach can correctly diagnose inaccurate counting introduced by suboptimal reaction conditions. For instance, the Sandberg lab team explained that their new tool revealed “completely artificial” results from a variant of the SCRB-seq protocol. This effect is caused by preferential priming of UMI-containing oligo-dT primers during PCR amplification, thus introducing false UMIs every PCR cycle and resulting in an overcount of those UMIs and a false read.
To further validate the spike-UMI approach, the authors then took advantage of the F.SIGHT OMICS and its unique feature to capture nozzle brightfield images for each isolated single cell along with the corresponding cell diameter. Single HEK293FT cells were precisely deposited into 384-well plates containing only 0.3 μL lysis buffer (+3 μL hydrophobic overlay) and then processed according to the Smart-seq3xpress protocol. By calculating the ratio of spike-in molecules to the cells’ endogenous RNA molecules, the authors could infer the absolute mRNA content of each cell which correlated to its diameter (Figure 1). The clear is normally masked by varying levels of sequencing saturation, which could successfully be corrected using the spike-in approach.

CONCLUSION
The study clearly showed that the molecular spikes can be applied in scRNA-seq experiments to diagnose and quantify artifactual RNA counting. Moreover, the authors used the F.SIGHT OMICS to truly play out the unique strengths of the device: highly precise, fast and efficient single-cell dispensing, and the simultaneous recording of true image data on each single cell dispensed. Using this comprehensive image data, researchers can integrate cell diameter in their analysis of scRNA-seq datasets, contributing to the complexity, accuracy and reliability of the results.
References:
Ziegenhain, C., Hendriks, GJ., Hagemann-Jensen, M. et al. Molecular spikes: a gold standard for single-cell RNA counting. Nat Methods 19, 560–566 (2022). https://doi.org/10.1038/s41592-022-01446-x
Products used:
F.SIGHT™ OMICS
Aim:
Capture nozzle brightfield images for each isolated single cell along with the corresponding cell diameter to validate the spike-UMI approach.






